PacVIEW Tutorial 1
Receptor-Ligand Interaction Visualisation Mode
This example will guide a new user through the graphic 3D visualisation of molecules using
PacVIEW - Receptor-Ligand Interaction Mode. For this tutorial, we will visualise and analyse the "re-docking" results of the complex between a minor groove binding drug, netropsin, and the DNA dodecamer d(CGCGATATCGCG) with a resolution of 2.4 Å (PDB code: 1DNE). The ligand (netropsin) has been separated from the complex structure of the target, and then docked back into the target using the docking program HYBRID (OpenEye). The docking program output is the best docked pose of the ligand. This file will be used as our input to visualise the ligand into the binding pocket of the receptor and the key receptor-ligand interactions.
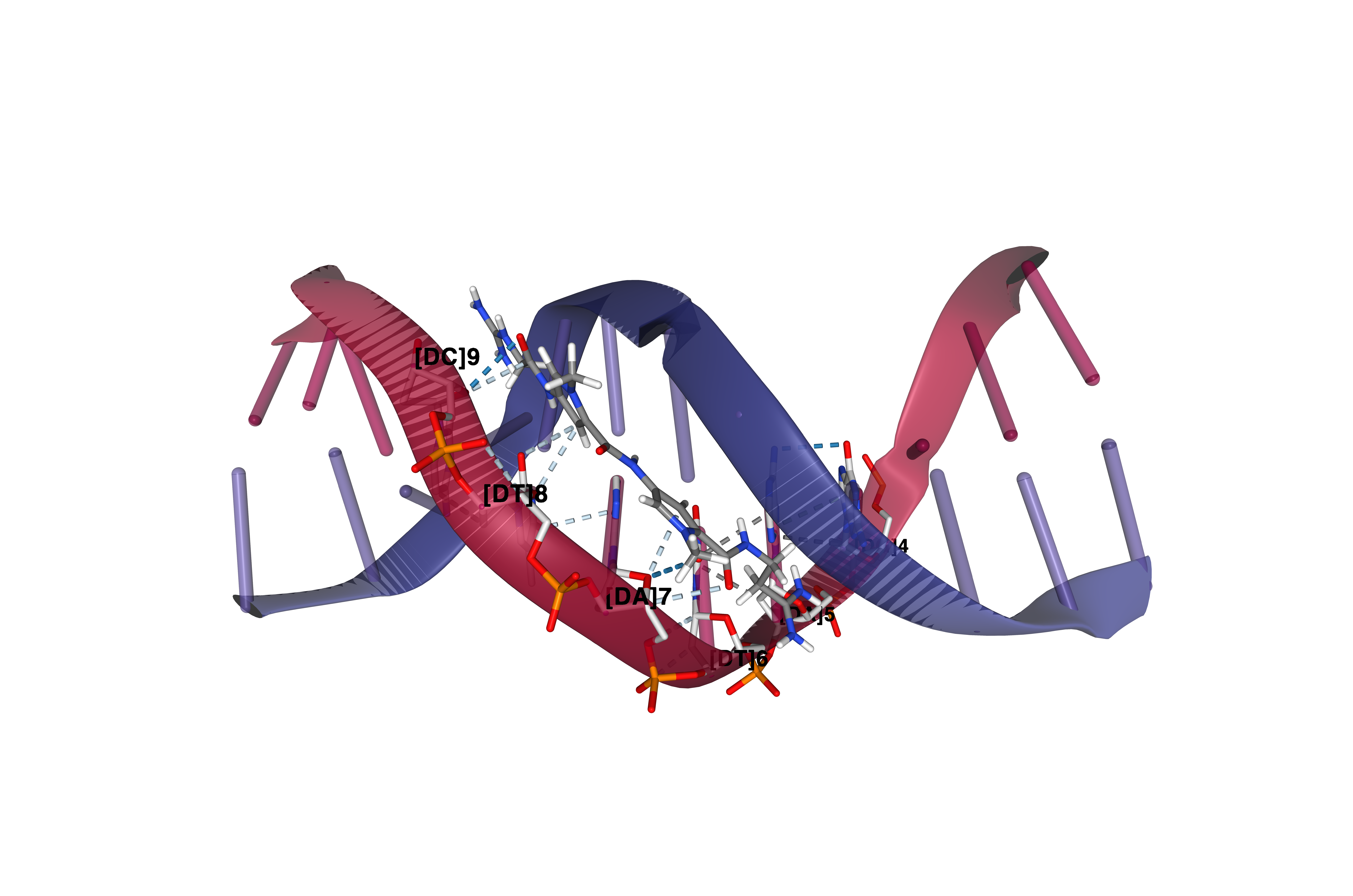
Interactions between the molcular structures of the DNA dodecamer-netropsin complex.
> For more information about PacVIEW, see PacVIEW Documentation.
PacVIEW - Receptor-Ligand Interaction Mode. For this tutorial, we will visualise and analyse the "re-docking" results of the complex between a minor groove binding drug, netropsin, and the DNA dodecamer d(CGCGATATCGCG) with a resolution of 2.4 Å (PDB code: 1DNE). The ligand (netropsin) has been separated from the complex structure of the target, and then docked back into the target using the docking program HYBRID (OpenEye). The docking program output is the best docked pose of the ligand. This file will be used as our input to visualise the ligand into the binding pocket of the receptor and the key receptor-ligand interactions.
- Download the input files: click on the two files to download them
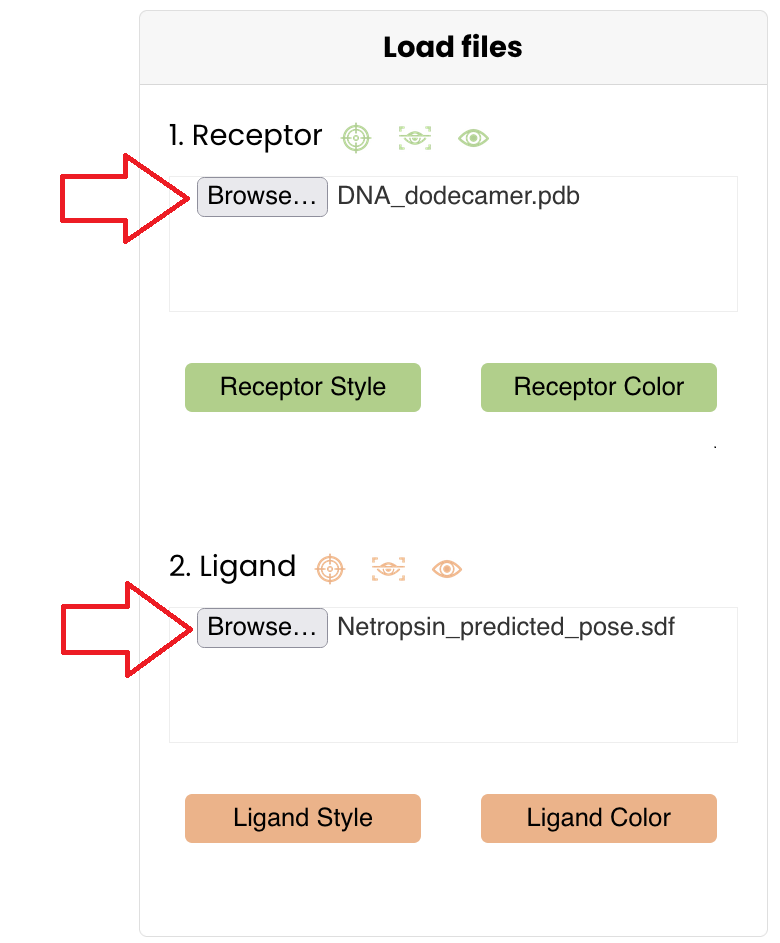
- Receptor structure: DNA_dodecamer.pdb
- Ligand structure: Netropsin_predicted_pose.sdf
-
Upload these files in the "Load files" section of the tool Click on "Browse..." and select them or drag them in their respective box
- Choose the Receptor Style and Receptor Color:
- the button allows you to choose the most appropiate style for the receptor (try Nucleic Acid)
- the button allows you to choose the most appropiate color for the receptor
: click this button to center the visualisation on the receptor - Choose the Ligand Style and Ligand Color:
- the button allows you to choose the most appropiate style for the ligand
- the button allows you to choose the most appropiate color for the ligand
: click this button to center the visualisation on the ligand -
Visualise the Receptor-Ligand Interactions: In this tutorial we will investigate the key receptor-ligand interactions, thus tick the box to display the Receptor-Ligand Interactions
You can choose the type of interactions to be investigated by switching their corresponding buttons. You can disable the residue labels turning-off the corresponding button. You can also choose label color by clicking the corresponding button and selecting one of the options.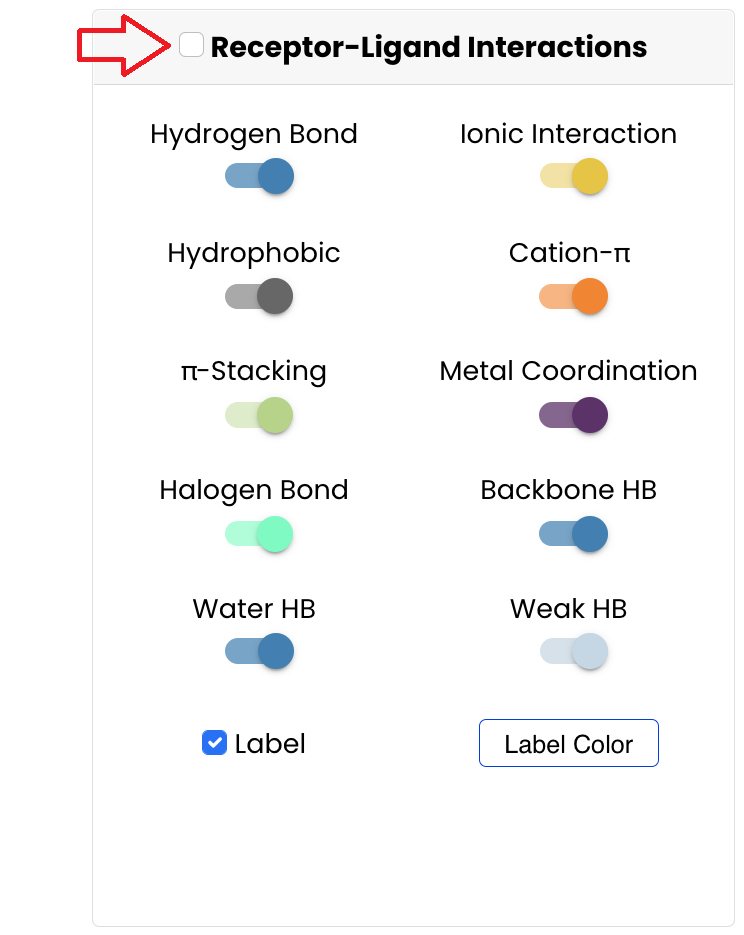
- Choose visualisation options:
- the Light Background switch allows you to change the background from black to white
- the Spin and Rock switches allow you to play two looping animation styles

- Click the button to save an image of the visualised molecules and interactions at different resolutions: Medium Quality and High Quality
- Click the button to enter the full screen mode (and "Esc" on your keyboard to exit the full screen mode)
- Click the button to center the visualisation on all molecules displayed
- Click the button to remove all molecules displayed
- the Receptor Opacity range slider allows you to opacify the receptor until hidden
- the Receptor Clip range slider allows you to clip the receptor

: click this button to hide the receptor
: click this button to display again the receptor
: click this button to hide the ligand
: click this button to display again the ligand


