ProRMSD Tutorial 1
a tool for automatic atom matching and RMSD calculation
This example will guide a new user through the calculation of the RMSD using ProRMSD.
For this tutorial, we will analyse the "re-docking" results of the cryo-electron microscopy structure of the μOR Opioid Receptor-Gi Protein Complex bound to the agonist peptide DAMGO and nucleotide-free Gi with a resolution of 3.5 Å (PDB code: 6DDF). DAMGO is a flexible opioid peptide, which has multiple rotatable bonds.
The ligand (DAMGO) has been separated from the complex structure of the target (μOR), and then docked back into the target using the docking program HYBRID (OpenEye).
The docking program output is composed of five poses of the ligand.
This file will be used as our input to calculate the RMSD of the predicted poses of the ligand from the known experimental pose in the protein-ligand complex.

> For more information about ProRMSD and how to interpret the results, see ProRMSD Documentation.
- Download the input files: click on the two files to download them
- Input file containing a molecular structure chosen as reference: DAMGO_experimental_pose.sdf
- Input file containing one or more molecular structures: DAMGO_predicted_poses.sdf
- Upload these files in the "Upload the structure files" section of the ProRMSD tool
Click on "Browse..." and select them or drag them in their respective box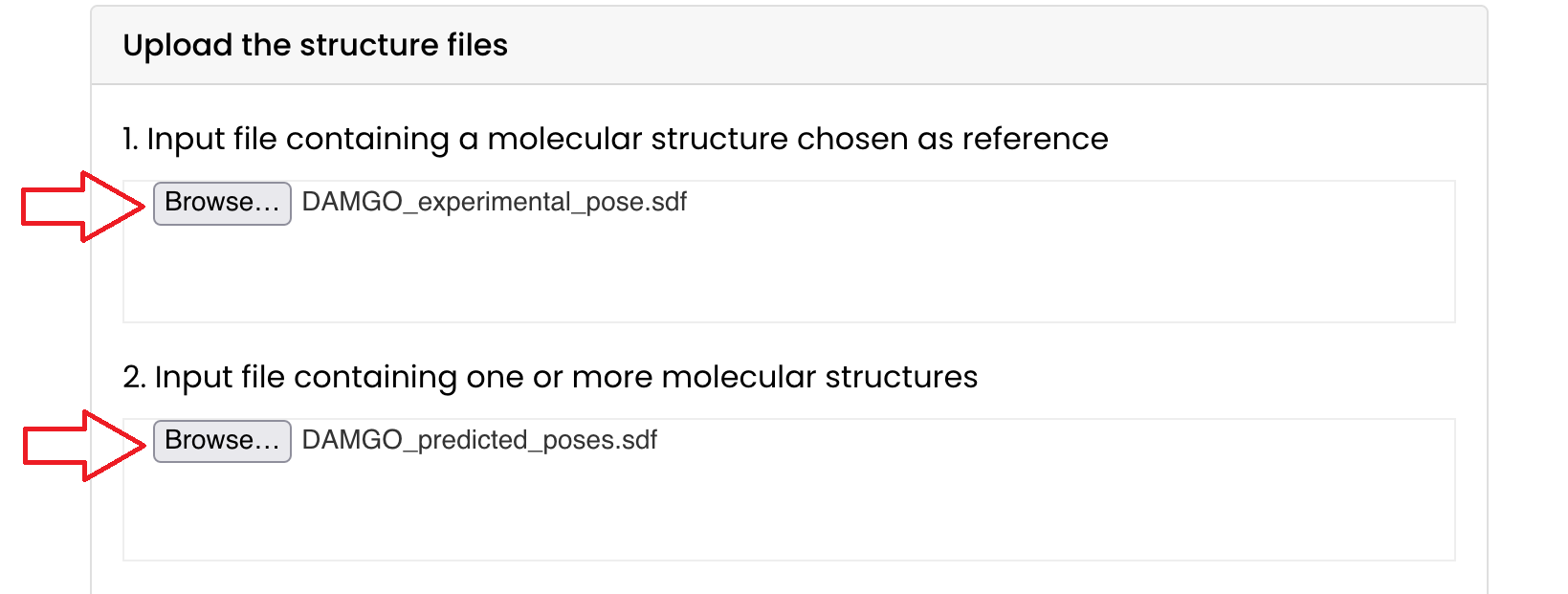
- Choose the calculation mode: in this tutorial we will calculate the RMSD with the standard algorithm, thus select the "RMSD" button
If you want to calculate the Backbone RMSD or the Superimposed RMSD click on their respective button
- Calculate the RMSD by clicking the "Calculate" button